News
-
Master 2 internship position available
2024-07-17
-
Welcome to Denez Le Collen, our Master's intern this semester.
2024-06-06
-
Our new article on histone phosphorylation and genome maintenance
2024-01-29
-
Openings for PhD students and postdocs
2024-01-29
-
Welcome to Lexin Xu, our new PhD student!
2024-01-29
-
Congratulations to Celia on her PhD defense!
2023-07-10
-
Lilian's paper on chromosomal organization and DNA replication
2022-10-25
-
The lab has moved to Bordeaux!
2022-10-25
The Wu Lab

Genome duplication is a critical part of the cell cycle that is highly regulated to ensure proper cell growth and proliferation. The distribution of the sites of initiation of DNA replication, or origins, across the genome changes during development and differentiation, suggesting that the program of genome duplication is highly controlled. Alterations in the replication pattern have also been observed in a number of pathologies, such as in cancers. However, the fundamental features driving origin selection and the importance of using specific replication programs remain surprisingly unknown. The research in our laboratory aims to study different aspects of genome duplication and maintenance using the fission yeast Schizosaccharomyces pombe as a model system.
- Background
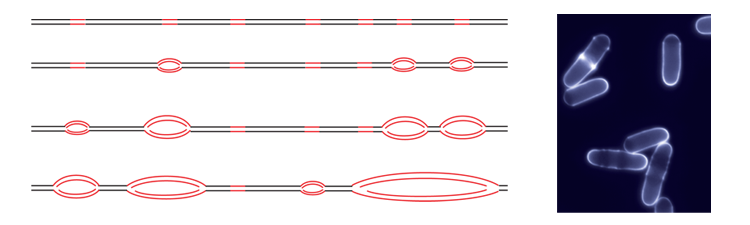
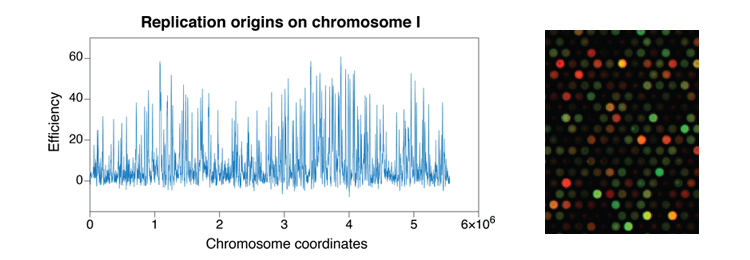
- Eukaryotic genome duplication initiates at sites called origins of replication distributed throughout the genome. The activity of an origin is defined by two major parameters: the time at which it fires during S phase and the efficiency, or probability, of its usage in a given cell. Origin usage has a stochastic component: each cell uses a subset of its origins during each cell cycle, and different origins are used in genetically identical cells grown in the same conditions. However, it is also clear that at the different cell types have different overall programs of origin usage. To achieve this, the mechanisms that control origin selection must integrate both stochastic and deterministic elements. In fission yeast, approximately 900 potential replication origins have been mapped across the three chromosomes. Unlike origins in budding yeast, which have a short and well-defined consensus sequence, S. pombe origins are more extended in length, have a high A-T content, and located in intergenic regions. These characteristics, reminiscent of more complex higher eukaryotic origins, make fission yeast an ideal model organism in which to investigate the control of the program of DNA replication.
- Establishment and organization of the replication program
- While there are a large number of parameters that have been shown to affect origin activity, ranging from DNA sequence to active transcription to chromatin modifications, it remains unknown how all of these inputs work together to determine the overall program of replication. We are developing novel approaches to investigate the core parameters that organize origin selection. In particular, we take advantage of a situation in which cells exit the cell cycle and then re-establish the replication program de novo. This approach will allow us to gain insight into the first critical steps in origin selection.
- Cyclin-dependent kinase control of replication origin selection
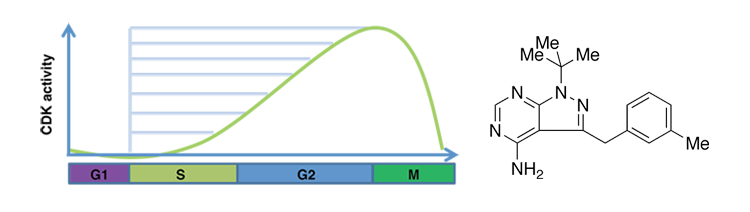
- Despite the critical function of the conserved cell cycle cyclin-dependent kinase (CDK) Cdc2 in S phase entry and checkpoint regulation, surprisingly little is know about CDKs affect origin selection and the overall program of replication. Using a strain of S. pombe that allows us to precisely modulate CDK activity using chemical genetic methods at any point during the cell cycle, we aim to uncover the quantitative relationship between CDK levels and origin usage.
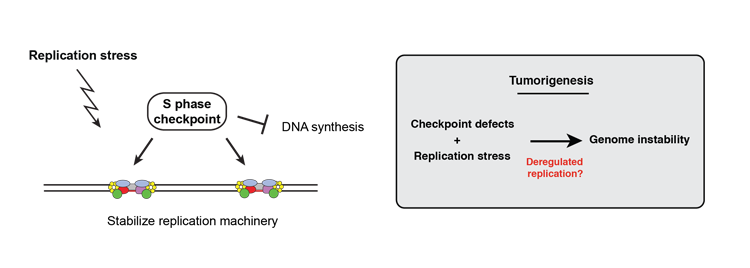
- Impact of the program of DNA replication on genome integrity
- We are interested in understanding how global changes in the replication program have consequences on cellular functions, and particularly how a de-regulation of the replication program can have consequences on genome stability. Our initial observations indicate that checkpoint defective cells subjected to replication stress show altered replication patterns. This mimics the situation encountered in the early steps of cell transformation, where checkpoint defects and changes in the pattern of replication are frequently observed. Since the link between alterations in origin firing and genome instability is difficult to uncover in cancer cells due to the complexity and heterogeneity of tumors, we are using fission yeast as a simple model to decipher how mutations may arise during the early steps of cancerogenesis and contribute to genome instability.
- Functional importance of the organization of DNA replication
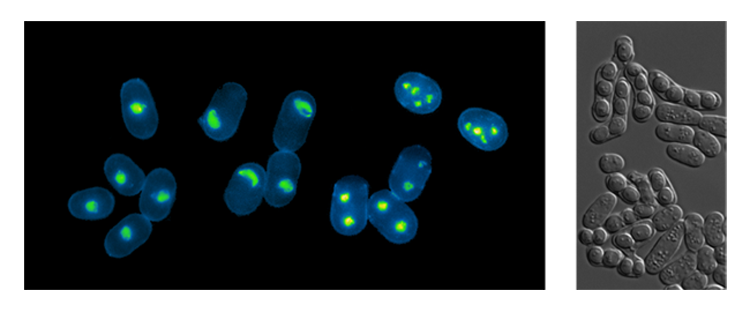
- While it is clear that the temporal and spatial organization of DNA replication is a conserved feature in eukaryotic organisms, the importance of this regulation remains largely unexplored. Indeed, although changes in this pattern of replication have been observed during development and in pathological situations, it remains unknown whether undergoing S phase with particular programs of origin selection has important consequences on cellular function. We have addressed this question using the process of meiosis in the fission yeast as a model system. Our results have demonstrated that genome-wide alteration of the replication program of pre-meiotic S phase does not affect overall meiotic progression or spore viability, indicating that meiosis does not intrinsically activate or critically require a particular pattern of origin usage, in contrast to previous models. Strikingly, our findings revealed that origin selection is a major determinant for organizing meiotic recombination, providing the first evidence the genome may be shaped by the way it is replicated. Building on these results, we are using a combination of genome-wide and single-cell stuidies understand how the organization of DNA replication contributes to the generation of genetic diversity through meiotic recombination.
- Selected publications
- Equipment
- Resources & Links
- See all publications
| The organization of genome duplication is a critical determinant of the landscape of genome maintenance Gomez-Escoda B and Wu PY Genome Research 28:1179-1192 (August 2018) Abstract Download PDF |
| CDK activity provides temporal and quantitative cues for organizing genome duplication Perrot A, Millington CL, Gomez-Escoda B, Schausi-Tiffoche D, and Wu PY PLOS Genetics 14(2):e1007214 (February 2018) Abstract Download PDF |
-
Equipment for working with yeast:
Innova 3100: High-precision temperature-controlled water bath incubators for growing liquid yeast cultures.
Memmert incubators: Temperature-controlled incubators for growing yeast at varying temperatures (priamrily 25ºC, 32ºC, or 36ºC).
Zeiss Axiovert microscopes: Two simple inverted microscopes for rapid visualization of cells growing in liquid media and on plates.
Dissection microscopes: Two Singer microscopes (MSM-2000 and Sporeplay) designed for the micromanipulation of yeast spores.
-
Genome-wide analysis of DNA replication:
Fastprep (MP Biochemicals): Benchtop cell disruptor for the lysis of biological samples, thoroughly lyses yeast cells for the isolation of genomic DNA, proteins, and RNA.
Bioruptor (Diagenode): Sonication system that uses a water bath to generate ultrasound waves, fragmenting DNA or chromatin.
SpeedVac (ThermoFisher): For concentration of samples by evaporation using vacuum and elevated temperatures.
Hybridization oven (Agilent): For optimal hybridization of microarray experiments, with adjustable temperatures and speeds.
-
Flow cytometry:
Accuri C6 (Becton Dickinson): An easy-to-use flow cytometer that is simple to maintain. The system is equipped with a blue and a red laser, two light scatter detectors, and four fluorescence detectors with optical filters optimized for the detection of fluorochromes such as propidium iodide, Sytox Green, and GFP, among others.
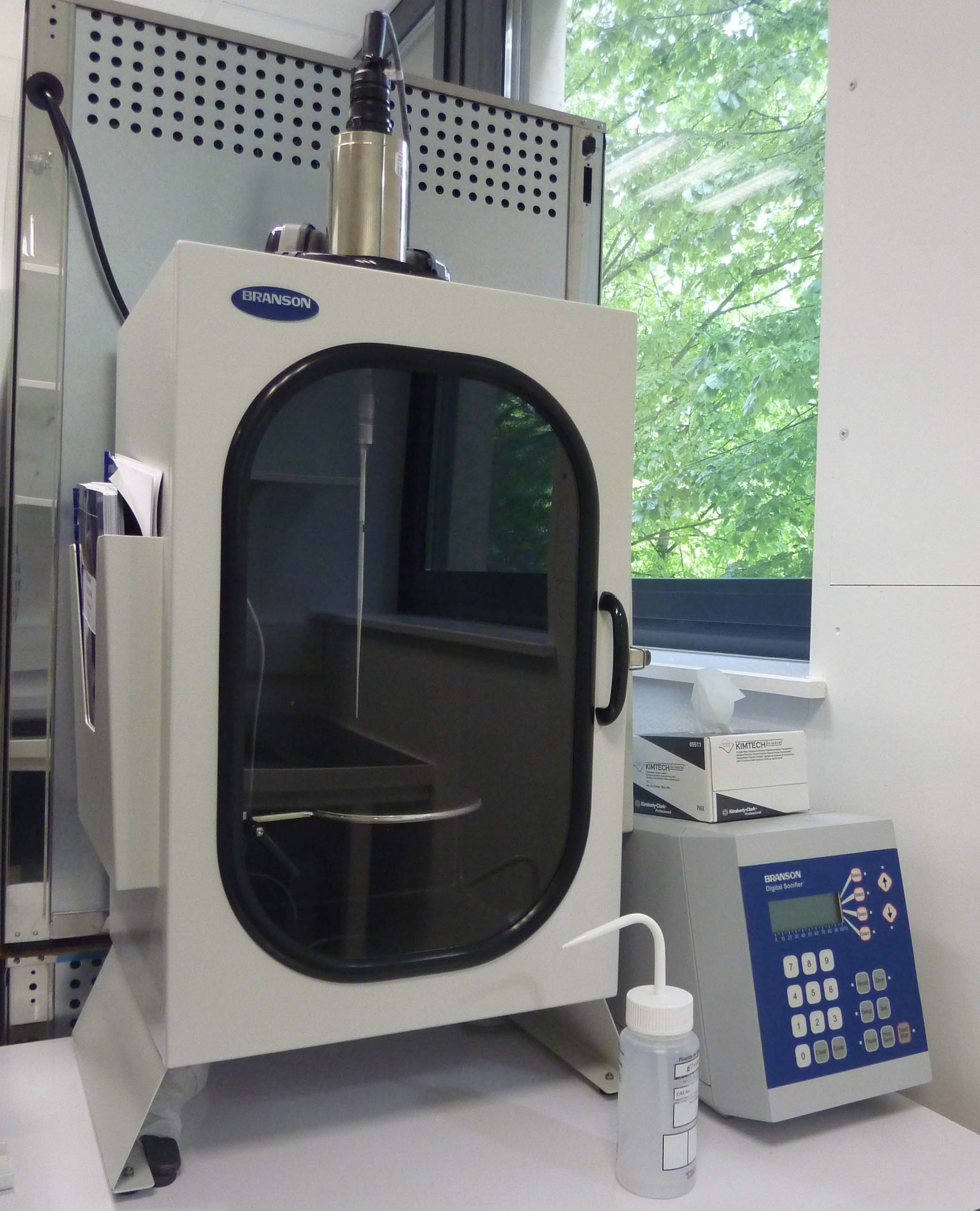
Sonicator (Branson): Generates ultrasonic vibrations for cell disruption, chromatin shearing, or yeast cell separation for flow cytometry.
-
Single-molecule analysis of DNA replication:
Molecular Combing system (Genomic Vision): For the direct visualization and analysis of single DNA molecules, where DNA is stretched and attached to a specially-treated glass surface. In our studies, sites of DNA synthesis can be labelled in vivo and then analysed by DNA combing to explore the organization of genome duplication along individual chromosomes. -
Imaging:
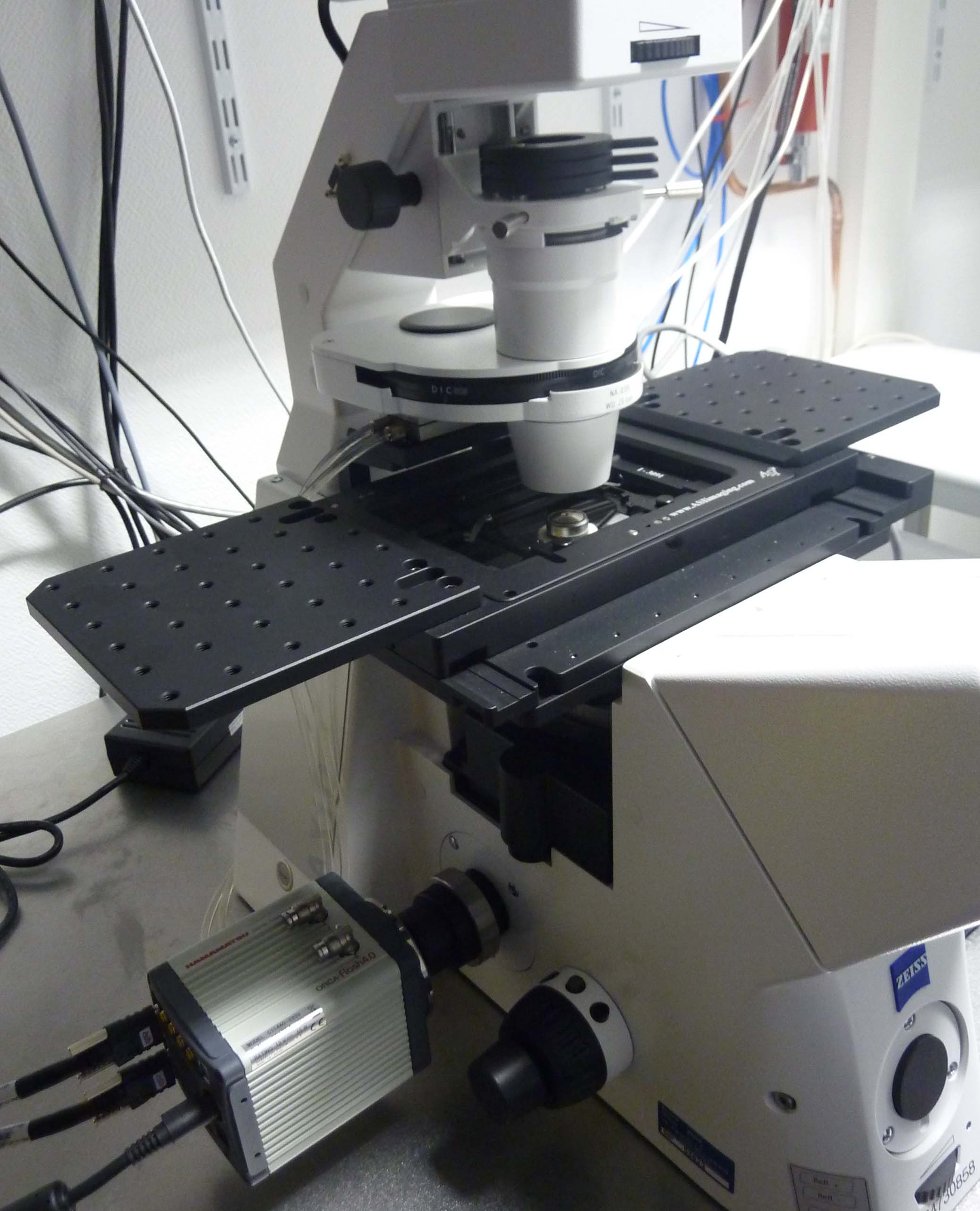
Axio Observer Z1 microscope (Zeiss): Inverted microscope equipped with LED illumination for epifluorescence (Lumencor Spectra X) and a sCMOS camera (Hamamatsu ORCA Flash 4.0 V2). This setup is fully motorized, with an ASI PZ-2000FT XY-Z piezoelectric stage, and it is coupled with flow and temperature control systems for live-imaging of cells grown in microfluidic chambers. Image acquisition is controlled by the VisiView package from Visitron Systems GmbH.













- Français
- English